Fabrication of Protein Microarrays
Background. The accurate and rapid detection of illegal
drugs are two of the principal goals of forensic drug analysis. Until recently,
only two types of conventional detection techniques were widely used:
immunoassays and chromatography.1-6 Immunoassay-based techniques (e.g., enzyme multiple immunoassay test (EMIT),
radio immunoassay (RIA), fluorescence polarization immunoassay (FPI)) make use
of immunochemical reactions triggered by antigen-antibody
interactions to detect illegal substances and they are primarily used in an
initial screening. Antibodies that bind selectively and specifically to target
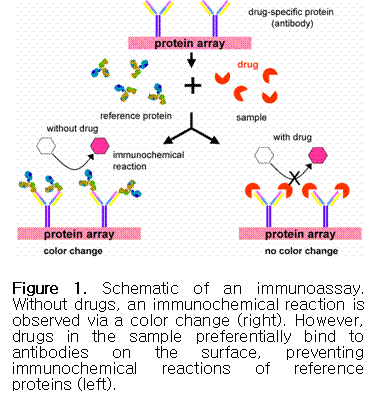
drugs (or drug metabolites) are immobilized on the surface of test kits. If
target compounds exist in the sample, they will preferentially bind to
corresponding antibodies on the test kit, preventing
immunochemical reactions (see Figure 1). The amount of drug in the
sample can then be detected through dye-labeled reference enzymes, radioisotopes,
or fluorescent compounds. Despite their rapid and on-site detection, false positive results are frequently observed
in immunoassays primarily because of low sensitivity and selectivity toward
target drug compounds.
 On the other hand,
chromatography-based tests (e.g., GC/MS) are well-established; they are used to
confirm test results if drugs are found in the first screening step. Although these techniques are capable of simultaneously
determining the presence of a wide variety of different compounds, on-site
and on-time access to these techniques is limited due to their
restricted portability and availability. This fact calls for the
development of novel portable devices that can be used for the in situ detection and identification of
illegal drugs with enhanced sensitivity and selectivity.
On the other hand,
chromatography-based tests (e.g., GC/MS) are well-established; they are used to
confirm test results if drugs are found in the first screening step. Although these techniques are capable of simultaneously
determining the presence of a wide variety of different compounds, on-site
and on-time access to these techniques is limited due to their
restricted portability and availability. This fact calls for the
development of novel portable devices that can be used for the in situ detection and identification of
illegal drugs with enhanced sensitivity and selectivity.
Miniaturized and parallelized array-based
devices are of great significance due to their ability to simultaneously
identify multiple target compounds within a single experiment with obvious applications
in forensics and public health.7-11 In
this proposal, the preparation and evaluation of microsized
protein arrays for the detection of drugs of abuse will be described. Compared
to conventional preparation techniques, two new strategies will be developed:
incorporation of a concentration gradient along the arrays for quantitative
drug analysis and the control of protein orientation via imine
coupling.12-14
Proposed Experiments. Protein array-based biosensors can be fabricated in two steps: (a) preparation of functionalized and patterned surfaces as templates for site-selective protein immobilization (e.g., antibodies for corresponding drugs of abuse) and (b) subsequent protein immobilization on the designated patterned areas. One of the ways to introduce functionalities on the solid surface for controlled protein adsorption can be achieved by the use of self-assembled monolayers (SAMs).9,11,15-17 Typical SAMs can be formed on gold and silica surfaces using thiols (X-(CH2)n-SH, X = CH3, OH, CF3, NH2, etc) and silanes (X-(CH2)n-SiCl3, X = CH3, OH, CF3, NH2, etc), respectively. SAMs formed in each patterned area introduce various chemical and physical properties on solid substrates, which act as platforms for selective and specific protein immobilization.
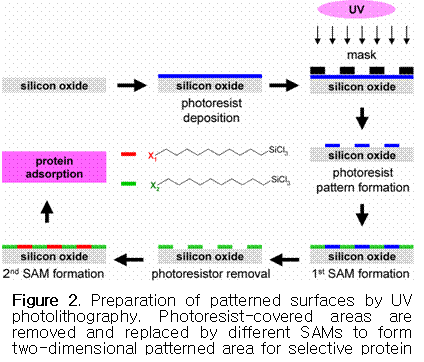 In the case of silicon oxide surfaces (silica or glass), a
general procedure for patterning by photolithography is illustrated in Figure 2.18-21 First, a photoresist thin film
is spin-cast on the silicon oxide surface. Photoresist
patterns are formed on the silicon oxide by UV illumination through a mask. The
photoresist in the UV-illuminated areas is removed
and filled in by a SAM of the silane with a terminal
group X1 (e.g., N-hydroxysuccinimide,
NHS). The remaining photoresist is then removed by UV
radiation followed by the formation of a SAM using a second silane
with a different terminal group X2
(e.g., hexaethylene glycol, EG6OH). The silicon oxide surface is now covered with two
different SAMs to form patterns that have different
affinities toward proteins. A NHS-terminated SAM has been popularly used
to covalently immobilize proteins on the solid surface via the formation of
amide bonds between NHS ester moieties on the surface and primary amine groups
in proteins.22,23 On the other hand, an
EG6OH-terminated SAM has been used to passivate
surfaces toward nonspecific protein adsorption.24-26
In the case of silicon oxide surfaces (silica or glass), a
general procedure for patterning by photolithography is illustrated in Figure 2.18-21 First, a photoresist thin film
is spin-cast on the silicon oxide surface. Photoresist
patterns are formed on the silicon oxide by UV illumination through a mask. The
photoresist in the UV-illuminated areas is removed
and filled in by a SAM of the silane with a terminal
group X1 (e.g., N-hydroxysuccinimide,
NHS). The remaining photoresist is then removed by UV
radiation followed by the formation of a SAM using a second silane
with a different terminal group X2
(e.g., hexaethylene glycol, EG6OH). The silicon oxide surface is now covered with two
different SAMs to form patterns that have different
affinities toward proteins. A NHS-terminated SAM has been popularly used
to covalently immobilize proteins on the solid surface via the formation of
amide bonds between NHS ester moieties on the surface and primary amine groups
in proteins.22,23 On the other hand, an
EG6OH-terminated SAM has been used to passivate
surfaces toward nonspecific protein adsorption.24-26
Alternatively, patterned surfaces will be
prepared by microcontact printing on gold surfaces
using thiol molecules with different terminal groups.27-29 Unlike photolithography,
patterned surfaces will be formed by direct physical transfer of thiol molecules to designated areas by the use of stamps.
Stamps for this purpose will be made via in
situ polymerization of polydimethylsiloxane
(PDMS) on the dye. PDMS is popularly used because of its inert properties
toward thiol molecules, low cost, and easy handling.
Once thiols are transferred on gold by stamping, a
SAM is formed in short time (typically within 10 to 20 min). The remaining
areas will be filled with a second thiol to form a
SAM with a different terminal group. Figure 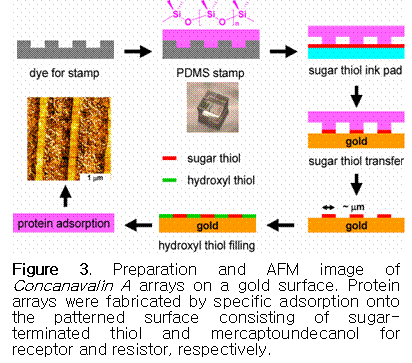 3 shows the procedure
to make a Concanavalin A pattern by microcontract printing using sugar- and hydroxyl-terminated
thiols on the gold surface. Concanavalin
A is selectively adsorbed on the area covered with sugar-terminated thiol via specific interactions.30
3 shows the procedure
to make a Concanavalin A pattern by microcontract printing using sugar- and hydroxyl-terminated
thiols on the gold surface. Concanavalin
A is selectively adsorbed on the area covered with sugar-terminated thiol via specific interactions.30
In both cases, surface gradients will be
introduced by controlling the residential time of bare gold (or silica)
surfaces in solutions with corresponding thiols (or silanes).31,32 By incorporating these surface concentration gradients,
arrays on the surface will have different concentrations of surface reactive
groups, allowing them to recruit different amounts of proteins in the next
step. Such gradient arrays can be used to determine the detection limit and
concentration of drugs in the sample.
Once pattered
surfaces are prepared, antibodies will be immobilized to localized areas
with no (or less) adsorption on the other area via non-specific physisorption,
specific physisorption (protein-ligand
interaction), or chemical coupling. This is critically important for immunoassays since the
optimum orientation of biologically active proteins immobilized on the surface
can greatly increase the surface binding ability and, therefore, sensitivity
toward target compounds. For more controlled
and stable protein adsorption, protein immobilization will be carried out by imine coupling between aldehyde
moieties in antibodies and hydrazido groups on the
surface.12-14 Aldehyde groups in antibodies can
be formed by the mild oxidation of carbohydrate moieties in antibodies using
sodium metaperiodate (NaIO4); hydrazido groups (-NH-NH2) are formed by the
reaction of surface NHS groups and hydrazine (H2NNH2).
Finally and most importantly,
protein patterns fabricated in this fashion will be evaluated by in vitro drug tests via immunochemical
reactions. By measuring the color change triggered by immunochemical reactions
toward reference samples, the sensitivity and selectivity of protein-array
based sensors will be evaluated and compared to that of commercially available
test kits.
References
(1) Jungreis, E. Spot Test Analysis, Clinical, Environmental, Forensic, and Geochemical Applications; second ed.; John Wiley & Sons, Inc.: New York, 1997.
(2) Immunoassay, A Practical Guide; Law, B., Ed.; Taylor and Francis Ltd.: London, UK, 1996.
(3) Handbook Of Forensic Drug Analysis; Smith, F. P.; Siegel, J. A., Eds.; Academic Press: St. Louis, 2004.
(4) DNA in Forensic Science: Theory, Techniques and Applications; Ross, A. M.; Robertson, J.; Burgoyne, L., Eds.; CRC Press: New York, 1990.
(5) Gas Chromatography in Forensic Science; Tebbett, I., Ed.; Ellis Horwood: New York, 1992.
(6) The Analysis of Drugs of Abuse; Cole, M. D.; Caddy, B., Eds.; CRC Press: New York, 1994.
(7) Biosensor principles and applications; Blum, L. J.; Coulet, P. R., Eds.; M. Dekker: New York, 1991; Vol. 14.
(8) MacBeath, G.; Schreiber, S. L. Science 2000, 289, 1760-1763.
(9) Blawas, A. S.; Reichert, W. M. Biomaterials 1998, 19, 595-609.
(10) Stears, R. L.; Martinsky, T.; Schena, M. Nature Medicine 2003, 9, 140-145.
(11) Smith, R. K.; Lewis, P. A.; Weiss, P. S. Prog. Surf. Sci. 2004, 75, 1-68.
(12) Hirota, J.; Michikawa, T.; Natsume, T.; Furuichi, T.; Mikoshiba, K. FEBS Lett. 1999, 456, 322-326.
(13) O'shannessy, D. J.; Wilchek, M. Anal. Biochem. 1990, 191, 1-8.
(14) Weiping, Q.; Bin, X.; Xu, B.; Yao, D. F.; Lin, Y. H.; Wu, L.; Wang, C. X.; Yu, F.; Lu, Z. H.; Wei, Y. Mater. Sci. Engr. C 1999, 8-9, 475-480.
(15) Ferretti, S.; Paynter, S.; Russell, D. A.; Sapsford, K. E. Trends Anal. Chem. 2000, 19, 530-540.
(16) Netzer, L.; Sagiv, J. J. Am. Chem. Soc. 1983, 105, 674-676.
(17) Sagiv, J. J. Am. Chem. Soc. 1980, 102, 92-98.
(18) Lee, K. N.; Shin, D. S.; Lee, Y. S.; Kim, Y. K. J. Micromech. Microeng. 2003, 13, 18-25.
(19) Revzin, A.; Russell, R. J.; Yadavalli, V. K.; Koh, W. G.; Deister, C.; Hile, D. D.; Mellott, M. B.; Pishko, M. V. Langmuir 2001, 17, 5440-5447.
(20) Brockman, J. M.; Frutos, A. G.; Corn, R. M. J. Am. Chem. Soc. 1999, 121, 8044-8051.
(21) Cremer, P. S.; Yang, T. J. Am. Chem. Soc. 1999, 121, 8130-8131.
(22) Patel, N.; Davies, M. C.; Hartshorne, M.; Heaton, R. J.; Roberts, C. J.; Tendler, S. J. B.; Williams, P. M. Langmuir 1997, 13, 6485 -6490.
(23) Jiang, K. Y.; Schadler, L. S.; Siegel, R. W.; Zhang, X. J.; Zhang, H. F.; Terrones, M. J. Mater. Chem. 2004, 14, 37-39.
(24) Feldman, K.; Hähner, G.; Spencer, N. D.; Harder, P.; Grunze, M. J. Am. Chem. Soc. 1999, 121, 10134-10141.
(25) Harder, P.; Grunze, M.; Dahint, R.; Whitesides, G. M.; Laibinis, P. E. J. Phys. Chem. B 1998, 102, 426-436.
(26) Herrwerth, S.; Eck, W.; Reinhardt, S.; Grunze, M. J. Am. Chem. Soc. 2003, 125, 9359-9366.
(27) Kane, R. S.; Takayama, S.; E., O.; Ingber, D. E.; Whitesides, G. M. Biomaterials 1999, 20, 2363-2376.
(28) Graber, D. J.; Zieziulewicz, T. J.; Lawrence, D. A.; Shain, W.; Turner, J. N. Langmuir 2003, 19, 5431-5434.
(29) Libioulle, L.; Beitsch, A.; Schmid, H.; Michel, B.; Delamarche, E. Langmuir 1999, 15, 300-304.
(30) Smith, E. A.; Thomas, W. D.; Kiessling, L. L.; Corn, R. M. J. Am. Chem. Soc. 2003, 125, 6140-6148.
(31) Morgenthaler, S.; Lee, S.; Zurcher, S.; Spencer, N. D. Langmuir 2003, 19, 10459-10462.
(32) Jeon, N. L.; Dertinger, K. W.; Chiu, D. T.; Choi, I. S.; Stroock, A. D.; Whitesides, G. M. Langmuir 2000, 16, A-F.